Health
New AI Tool Revolutionizes Lung Cancer Mutation Detection in Under an Hour
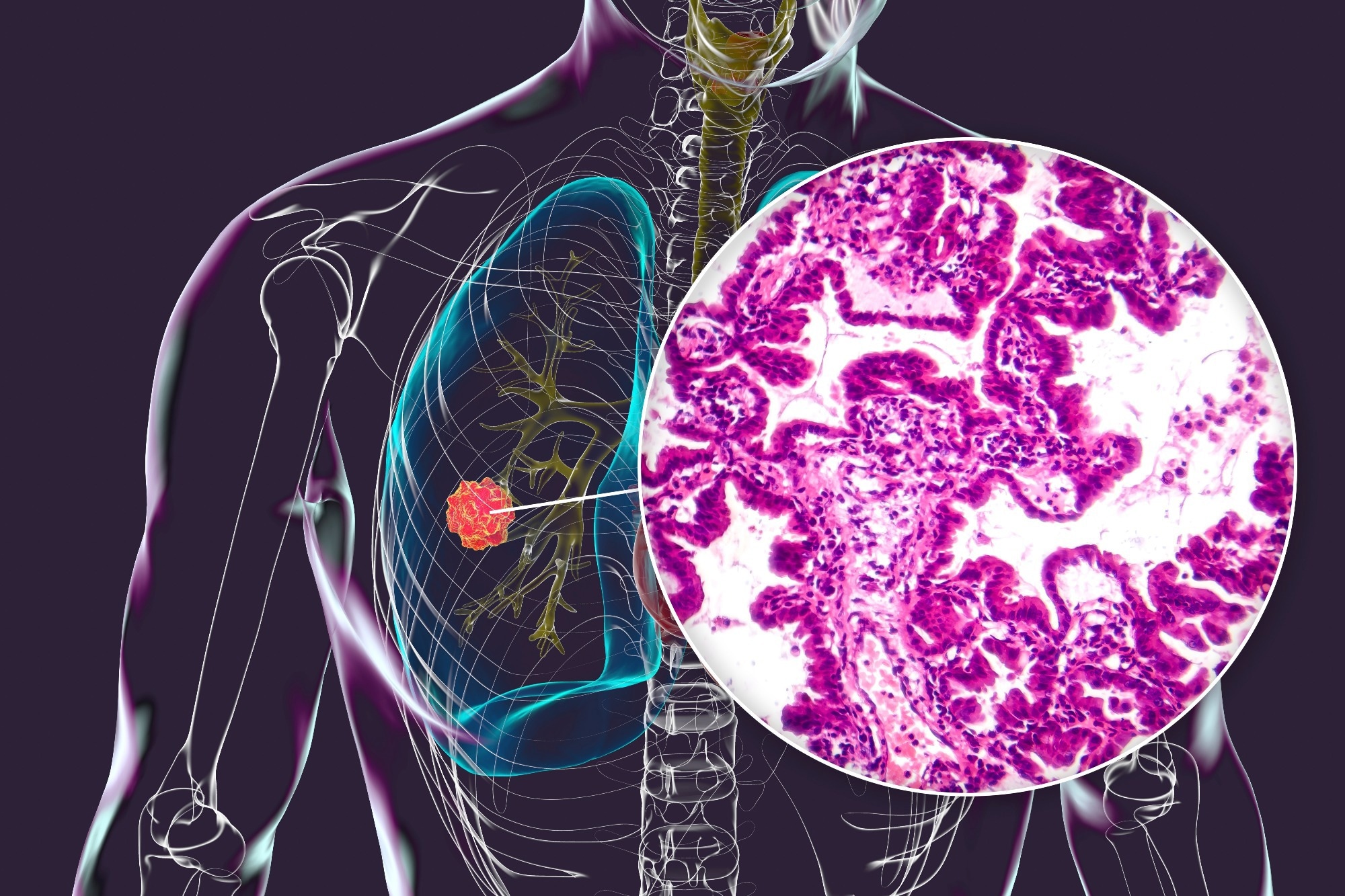
A groundbreaking artificial intelligence tool, known as EAGLE (EGFR AI Genomic Lung Evaluation), has been developed to analyze lung cancer biopsy slides in under an hour. This innovative approach accurately predicts mutations, thereby minimizing delays, reducing costs, and preserving valuable tissue samples. The findings represent a significant advancement in the field of precision cancer care.
In a study published in Nature Medicine, researchers compiled an extensive international clinical dataset of digital lung adenocarcinoma (LUAD) biopsy slides. The aim was to enhance the detection of the epidermal growth factor receptor (EGFR) biomarker, which is crucial for patients diagnosed with this prevalent form of lung cancer. Accurate EGFR testing is essential for ensuring that patients receive appropriate first-line therapies, particularly in advanced stages of LUAD.
Despite the importance of EGFR testing, it has been noted that testing rates fall short of established guidelines, often due to challenges in obtaining and processing biopsy samples. The EAGLE model addresses these issues by utilizing digitized pathology slides, specifically Hematoxylin and Eosin (H&E) biopsies, allowing for results to be reported without the need for physical processing. This efficiency not only streamlines the clinical workflow but also facilitates timely decision-making regarding patient treatment.
Study Design and Implementation
The study introduced EAGLE as a computational biomarker designed to enhance the existing molecular workflow. By leveraging diagnostic biopsies from LUAD patients, the model predicts EGFR mutational status while maintaining high-performance screening capabilities. Although traditional methods often delay results, the AI-assisted approach enables rapid testing, although next-generation sequencing (NGS) remains necessary for confirming positive screenings.
To train the EAGLE algorithm, researchers utilized a dataset comprising 5,174 biopsy slides from the Memorial Sloan Kettering Cancer Center (MSKCC). Validation of the model involved testing on 1,742 internal slides and multiple external cohorts across institutions in the United States and Europe. This rigorous validation process ensured that EAGLE’s performance was consistent across different scanners and clinical settings.
A silent trial was conducted to assess EAGLE’s real-world applicability. The model demonstrated expected performance on novel cases and was deemed suitable for clinical implementation. The comparison of EAGLE’s rapid test results with those of the Idylla test revealed a positive predictive value (PPV) of 0.988, sensitivity of 0.918, specificity of 0.993, and a negative predictive value (NPV) of 0.954.
Performance Insights and Future Directions
Despite strong overall performance, the model showed less accuracy in metastatic specimens, achieving an area under the curve (AUC) of 0.75 compared to 0.90 for primary samples. Notably, lymph node and bone samples yielded even lower AUCs of 0.74 and 0.71, respectively. These findings underscore the need for further refinement in detecting mutations in challenging specimen types.
The silent trial also explored threshold strategies to determine how many rapid tests could be eliminated while maintaining performance on par with traditional workflows. Depending on the selected threshold, the AI-assisted process reduced rapid tests by 18% to 43% without compromising the diagnostic accuracy.
One of EAGLE’s most significant advantages is its turnaround time, delivering results in a median of just 44 minutes. This is a stark contrast to the minimum 48 hours required for rapid tests and several weeks for NGS results. The silent trial provided valuable insights into the protocol’s performance and the identification of potential sources of false negatives and positives.
The researchers noted that false positives often involved biologically related mutations, while false negatives tended to arise in samples with minimal tumor architecture. This suggests that manual interpretation by pathologists could potentially reduce error rates significantly.
While EAGLE is not designed to replace NGS sequencing, it serves as an efficient screening tool to identify likely positive cases and rule out EGFR mutations. It is important to note that NGS confirmation remains essential before initiating treatment, as EAGLE does not differentiate between EGFR subtypes requiring distinct therapies.
The study highlights the promise of the EAGLE model in enhancing diagnostic efficiency and reducing tissue consumption in lung cancer testing. By fine-tuning a vision transformer-based foundation model, researchers are paving the way for more adaptable AI tools in pathology. Future research will focus on exploring additional biomarkers and conducting prospective clinical trials to further validate these findings.

-

 Health3 months ago
Health3 months agoNeurologist Warns Excessive Use of Supplements Can Harm Brain
-

 Health4 months ago
Health4 months agoFiona Phillips’ Husband Shares Heartfelt Update on Her Alzheimer’s Journey
-

 Science2 months ago
Science2 months agoBrian Cox Addresses Claims of Alien Probe in 3I/ATLAS Discovery
-

 Science2 months ago
Science2 months agoNASA Investigates Unusual Comet 3I/ATLAS; New Findings Emerge
-

 Science2 months ago
Science2 months agoScientists Examine 3I/ATLAS: Alien Artifact or Cosmic Oddity?
-

 Entertainment2 months ago
Entertainment2 months agoLewis Cope Addresses Accusations of Dance Training Advantage
-

 Entertainment5 months ago
Entertainment5 months agoKerry Katona Discusses Future Baby Plans and Brian McFadden’s Wedding
-
 Science2 months ago
Science2 months agoNASA Investigates Speedy Object 3I/ATLAS, Sparking Speculation
-

 Entertainment5 months ago
Entertainment5 months agoEmmerdale Faces Tension as Dylan and April’s Lives Hang in the Balance
-

 World3 months ago
World3 months agoCole Palmer’s Cryptic Message to Kobbie Mainoo Following Loan Talks
-

 World4 weeks ago
World4 weeks agoBailey and Rebecca Announce Heartbreaking Split After MAFS Reunion
-
 Science2 months ago
Science2 months agoNASA Scientists Explore Origins of 3I/ATLAS, a Fast-Moving Visitor